Abstract
In recent years, optimizing antibodies for biomedical applications has become increasingly important. However, traditional wet-experiment-based approaches are time-consuming and inefficient. To address this issue, we propose a diffusion model-based antibody optimization pipeline to improve binding affinity. Our approach involves two key models: AbDesign for designing antibody sequences and structures, and AbDock, a paratope-epitope docking model, used for scoring designed CDRs. On an independent test set, our AbDesign demonstrates the exceptional performance of an RMSD of 2.56Å in structure design and an amino acid recovery of 36.47% in sequence design. In a paratope-epitope docking test set, our AbDock achieves a state-of-the-art performance of DockQ 0.44, irms 2.71Å, fnat 0.40, and Lrms 6.29Å. The effectiveness of the optimization pipeline is further validated by experimentally optimizing a flaviviruse antibody 1G5.3, resulting in a broad-spectrum antibody that demonstrates improved binding to 6 out of the nine tested flaviviruses. This research offers a general-purpose methodology to enhance antibody functionality without training on data from specific antigens.
Method
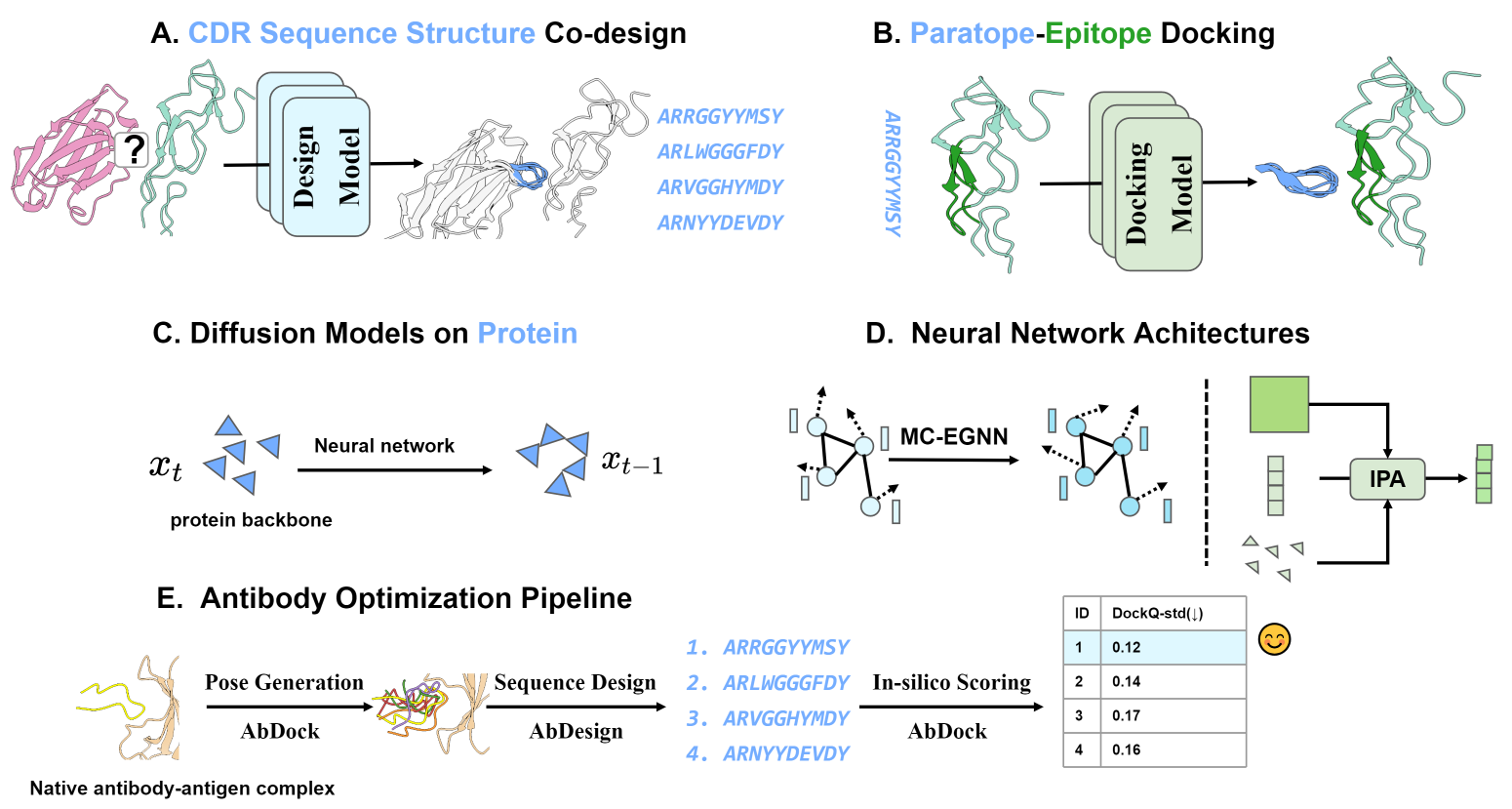
This study introduces two novel models, AbDesign and AbDock, aimed at antibody CDR sequence-structure co-design (A) and paratope-epitope docking (B), respectively. The co-design task involves predicting CDR sequence and conformation given a known paratope-epitope binding interface, while the docking task predicts paratope binding conformation when the epitope pocket and paratope sequence are known. Both models are built upon diffusion model foundations (C) and employ distinct neural network architectures based on the task requirements. The Multi-Channel Equivariant Neural Network (MC-EGNN) (left, D) is proposed for CDR sequence-structure co-design, effectively modeling contextual information of the binding interface. For paratope-epitope docking, the invariant point attention (IPA) network (right, D) is utilized to capture intricate structural complexities of the system. The study presents antibody optimization pipelines (E) using AbDesign for sequence design and AbDock for generating binding poses and in-silico scoring. |
 |
Results
Antibody CDRH3 Design
Animate the diffusion process of CDRH3 for PDB 5xku (left), 7chf_ABR (middle) and 7chf_HLR (right).
Visualize the designed CDRH3 of PDB 5xku (left), 7chf_ABR (middle) and 7chf_HLR (right).
Paratope-epitope Docking
Animate the the generation process of docking poses for PDB 5MES (left), 7BSD (middle) and 7DK2 (right).
Visualize the generated docking poses of PDB 5MES (left), 7BSD (middle) and 7DK2 (right).
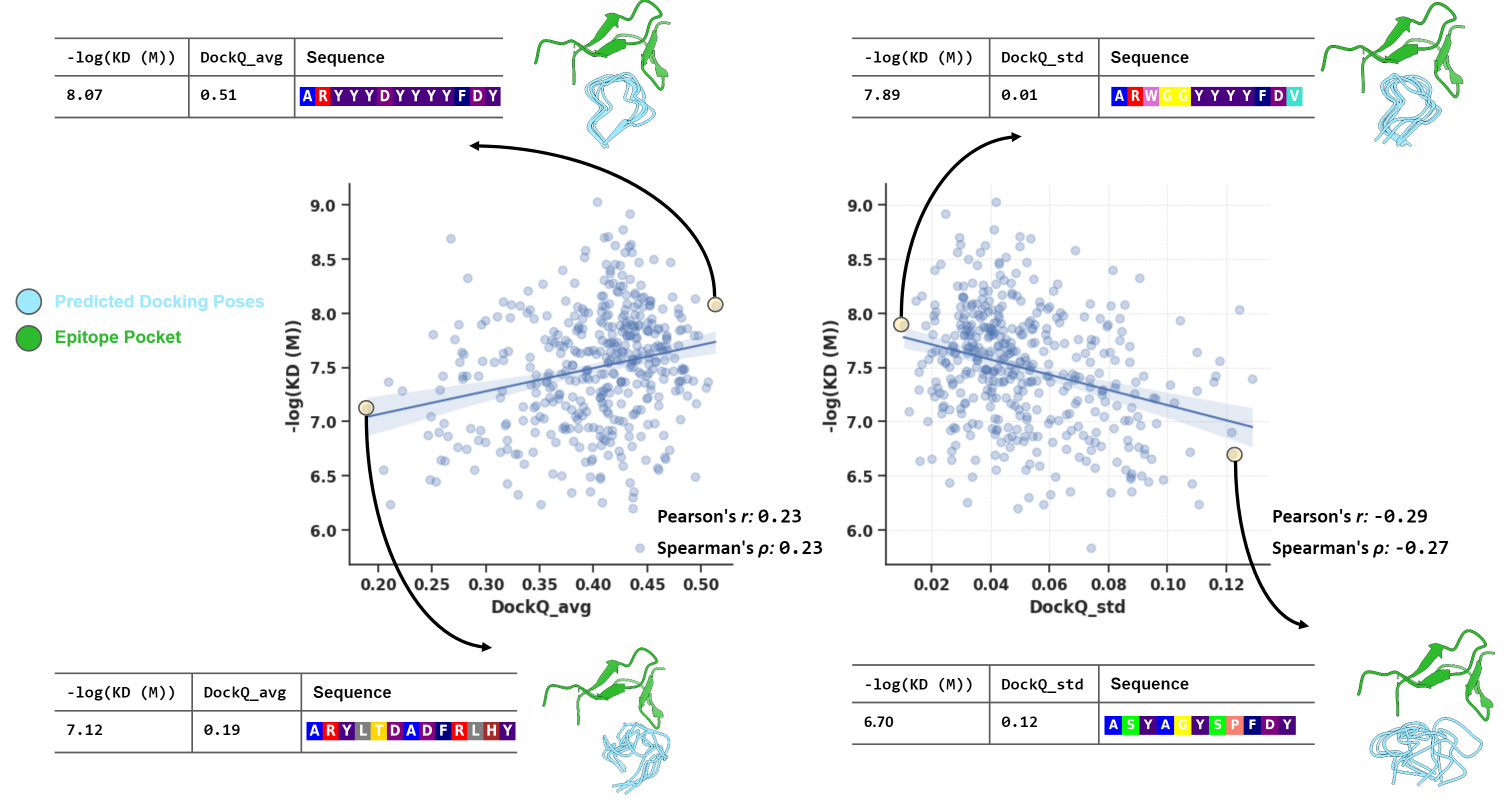
AbDock serves as a proxy for scoring designed CDRH3 sequences
Our AbDock, designed for paratope-epitope docking, can serve as a proxy for scoring designed CDRH3 sequences. We show the correlation between the AbDock score and the experimentally-determined affinity value for the designed CDRH3 sequences. |
 |
Wet experiments domostrate the effectiveness our antibody optimization
The effectiveness of the optimization pipeline is further validated by experimentally optimizing a flaviviruse antibody 1G5.3, resulting in a broad-spectrum antibody that demonstrates improved binding to 6 out of the 9 tested flaviviruses. |
 |